Note
Go to the end to download the full example code.
Analyze a post-training Bernoulli-Bernoulli RBM
This script shows how to analyze the RBM after having trained it.
import matplotlib.pyplot as plt
import torch
device = torch.device("cpu")
dtype = torch.float32
Load the dataset
We suppose the RBM was trained on the dummy.h5 dataset file, with 60% of the train dataset. By default, the dataset splitting is seeded. So just putting the same train_size and test_size ensures having the same split for analysis. This behaviour can be changed by setting a different value to the seed keyword.
from rbms.dataset import load_dataset
train_dataset, test_dataset = load_dataset(
"dummy.h5", train_size=0.6, test_size=0.4, device=device, dtype=dtype
)
num_visibles = train_dataset.get_num_visibles()
U_data, S_data, V_dataT = torch.linalg.svd(
train_dataset.data - train_dataset.data.mean(0)
)
proj_data = train_dataset.data @ V_dataT.mT / num_visibles**0.5
proj_data = proj_data.cpu().numpy()
Load the model.
First, we want to know which machines have been saved
from rbms.utils import get_saved_updates
filename = "RBM.h5"
saved_updates = get_saved_updates(filename=filename)
print(f"Saved updates: {saved_updates}")
Saved updates: [ 1 2 3 4 5 7 8 10 13 16 20 25
31 39 49 61 75 94 116 145 180 223 277 344
427 531 659 765 819 849 990 1017 1024 1262 1567 1568
1946 2118 2391 2416 2655 3000 3725 3799 4625 5743 7033 7131
8853 10992 13648 16946 18038 21040 26123 31967 32434 40270 42529 50000]
Now we will load the last saved model as well as the permanent chains during training Only the configurations associated to the last saved model have been saved for the permanent chains. We also get access to the hyperparameters of the RBM training as well as the time elapsed during the training.
from rbms.io import load_model
params, permanent_chains, training_time, hyperparameters = load_model(
filename=filename, index=saved_updates[-1], device=device, dtype=dtype
)
print(f"Training time: {training_time}")
for k in hyperparameters.keys():
print(f"{k} : {hyperparameters[k]}")
Training time: 4915.106016874313
batch_size : 2000
gibbs_steps : 5
learning_rate : 0.01
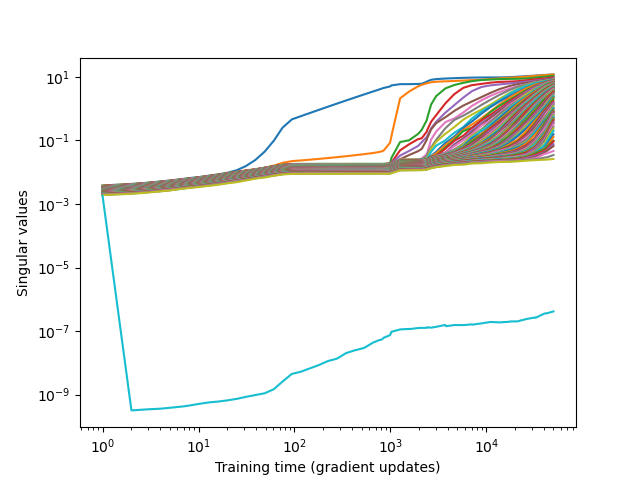
To follow the training of the RBM, let’s look at the singular values of the weight matrix
from rbms.utils import get_eigenvalues_history
grad_updates, sing_val = get_eigenvalues_history(filename=filename)
fig, ax = plt.subplots(1, 1)
ax.plot(grad_updates, sing_val)
ax.set_xlabel("Training time (gradient updates)")
ax.set_ylabel("Singular values")
ax.loglog()
fig.show()
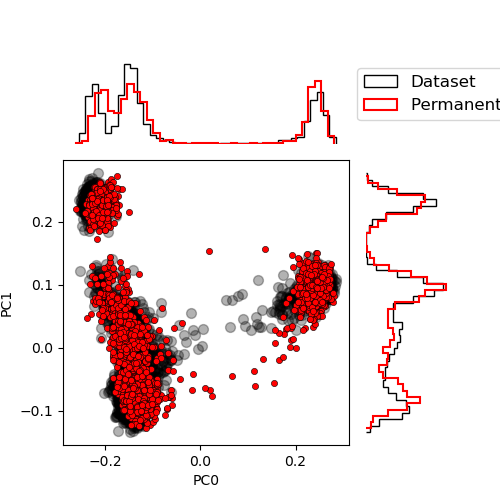
Let’s compare the permanent chains to the dataset distribution. To do so, we project the chains on the first principal components of the dataset.
from rbms.plot import plot_PCA
proj_pc = permanent_chains["visible"] @ V_dataT.mT / num_visibles**0.5
plot_PCA(
proj_data,
proj_pc.cpu().numpy(),
labels=["Dataset", "Permanent chains"],
)
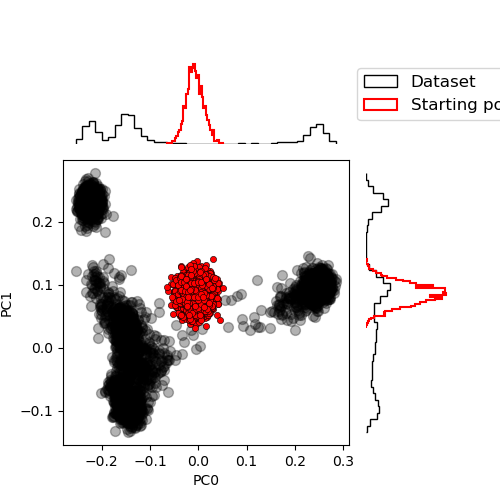
Sample the RBM
Another interesting thing is to compare generated samples starting from random configurations
from rbms.sampling.gibbs import sample_state
num_samples = 2000
chains = params.init_chains(num_samples=num_samples)
proj_gen_init = chains["visible"] @ V_dataT.mT / num_visibles**0.5
plot_PCA(
proj_data,
proj_gen_init.cpu().numpy(),
labels=["Dataset", "Starting position"],
)
plt.tight_layout()
/home/nbereux/work/rbms/examples/plot_generated_samples.py:100: UserWarning: Tight layout not applied. tight_layout cannot make Axes width small enough to accommodate all Axes decorations
plt.tight_layout()
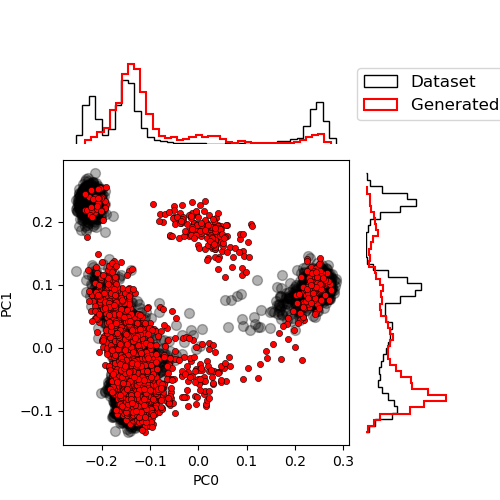
We can now sample those chains and compare again the distribution
n_steps = 100
chains = sample_state(gibbs_steps=n_steps, chains=chains, params=params)
proj_gen = chains["visible"] @ V_dataT.mT / num_visibles**0.5
plot_PCA(
proj_data,
proj_gen.cpu().numpy(),
labels=["Dataset", "Generated samples"],
)
Compute the AIS estimation of the log-likelihood.
For now, we only looked at a qualitative evaluation of the model
from rbms.partition_function.ais import compute_partition_function_ais
from rbms.utils import compute_log_likelihood
log_z_ais = compute_partition_function_ais(num_chains=2000, num_beta=100, params=params)
print(
compute_log_likelihood(
train_dataset.data, train_dataset.weights, params=params, log_z=log_z_ais
)
)
-387.64599609375
Total running time of the script: (0 minutes 8.271 seconds)